Often we log data in JSON, CSV or other text format to Amazon’s S3 as compressed files. This pattern is a) accessible and b) infinitely scalable by nature of being in S3 as common text files. However, there are some subtle but critical caveats that come with this pattern that can cause quite a bit of trouble. Here I’m going to discuss the “small files problem” that has existed in big data since Hadoop and MapReduce first came to prominence as well as advice on how to solve for it.
Consider the following spark command:
df = spark.read.json("s3://awesome-bucket/offers/2017/09/07/*")
That statement looks simple and innocuous enough. Tell spark to read in a day’s worth of JSON formatted files in awesome-bucket under the offers key “directory.”*
The “*” glob at the end of the path means we’ll be reading in the files in each hour ‘directory’, each of which contains over 30,000 files. In
total, there are 24 “directories” for a total of over 700,000 files and 72M rows, each file of which is ~30KB.
But… there’s a lot going on under the hood and it’s not good [for performance reasons].
1. S3 is not a file system
2. Meta data on tiny files
3. GZip compressed files
4. JSON
1. S3 is not a file system
Amazon’s Simple Storage Service (S3) is a “cloud-based object storage solution” where each ‘object’ is identified by a bucket and a key.
When you list a directory on your local computer (e.g. “ls /tmp” on *nix systems), the information returned about the directory and files is
returned immediately. Asking S3 to list all the files in a “directory” (hint: it’s not actually a directory), such as through “s3cmd ls s3://bucket
/path/to/files” returns results in at best seconds, possibly minutes. S3 is optimal when you have few large files but horrendous when you
have an army of tiny files, because more files means the listing process takes substantially longer.
2. Meta data on tiny files
Spark has to know exact path and how to open each and every file (e.g. s3://bucket/path/to/objects/object1.gz) even if you just pass a path
to a “directory” because ultimately that “directory” is just a collection of files (or “objects”). With an army of tiny files, this meta data gets
large, both in number of elements but also in terms of memory (100,000 records in a hashmap to denote location, compression type and
other meta data) is not lightweight. Add in the overhead of using S3 plus network latencies and it becomes more clear why this meta data
collection process takes a long time.
3. GZip Compressed Files
GZip compression is great. Sure there are other compression formats out there (e.g. brotli, lzo, bzip2, etc), few if any are as widespread and
accessible as GZip. But what GZip has in terms of market share, it sacrifices in more big data friendly features. GZip is not splittable, which
means the entire file must be processed by a single core/executor as a single partition. This is an anti-pattern in a tool like spark designed
explicitly around parallelism, because having to serially process a file is expensive. Small GZip files are actually better to process than larger
ones, because multiple cores can work on different small files at the same time and not sit idle as one or two cores do all the work. But
don’t be tricked into thinking “oh I’ll just have loads of small files” because as you saw above, loads of small files are far worse than just
about any alternative.
4. JSON
It’s slow; plain and simple. It allows for complex structures and data types as well as implicitly defined types, which all make it very
expensive to parse.
* Directory on S3 is a misnomer, because S3 is an object store where each object is a combination of a bucket (e.g. “awesome-bucket”) with
a specific key (e.g. “/path/to/objects/object1.gz”) that has the object’s contents as its value. Think of it like a key-value store where ”
s3://awesome-bucket/offers/2017/09/07/12/2017-09-07-12-59-0.9429827.gz” is the KEY and the gzipped contents of
that file are the value. The slashes after the bucket name (awesome-bucket) mean nothing to S3; they exist solely for the user’s
convenience. Treating them as if they denote true directories is a convenience feature Amazon and APIs offer.
What can we do about it?
Use file formats like Apache Parquet and ORC. If you need to work with (ideally converting in full) armies of small files, there are some
approaches you can use.
1) S3DistCP (Qubole calls it CloudDistCP)
2) Use scala with spark to take advantage of Scala and Spark’s unique parallel job submission. See below for an example.
3) Just wait. A long time.
Parallel Job Submission to Consolidate many “directories”
val hours = (0 to 23).map(h => "%02d".format(h)) // zero pad
hours.par.foreach(hour => {
spark.read.json("s3a://awesome-bucket/offers/2017/09/07/" + hour.toString +
"/*").write.repartition(16).parquet("s3://output")
})
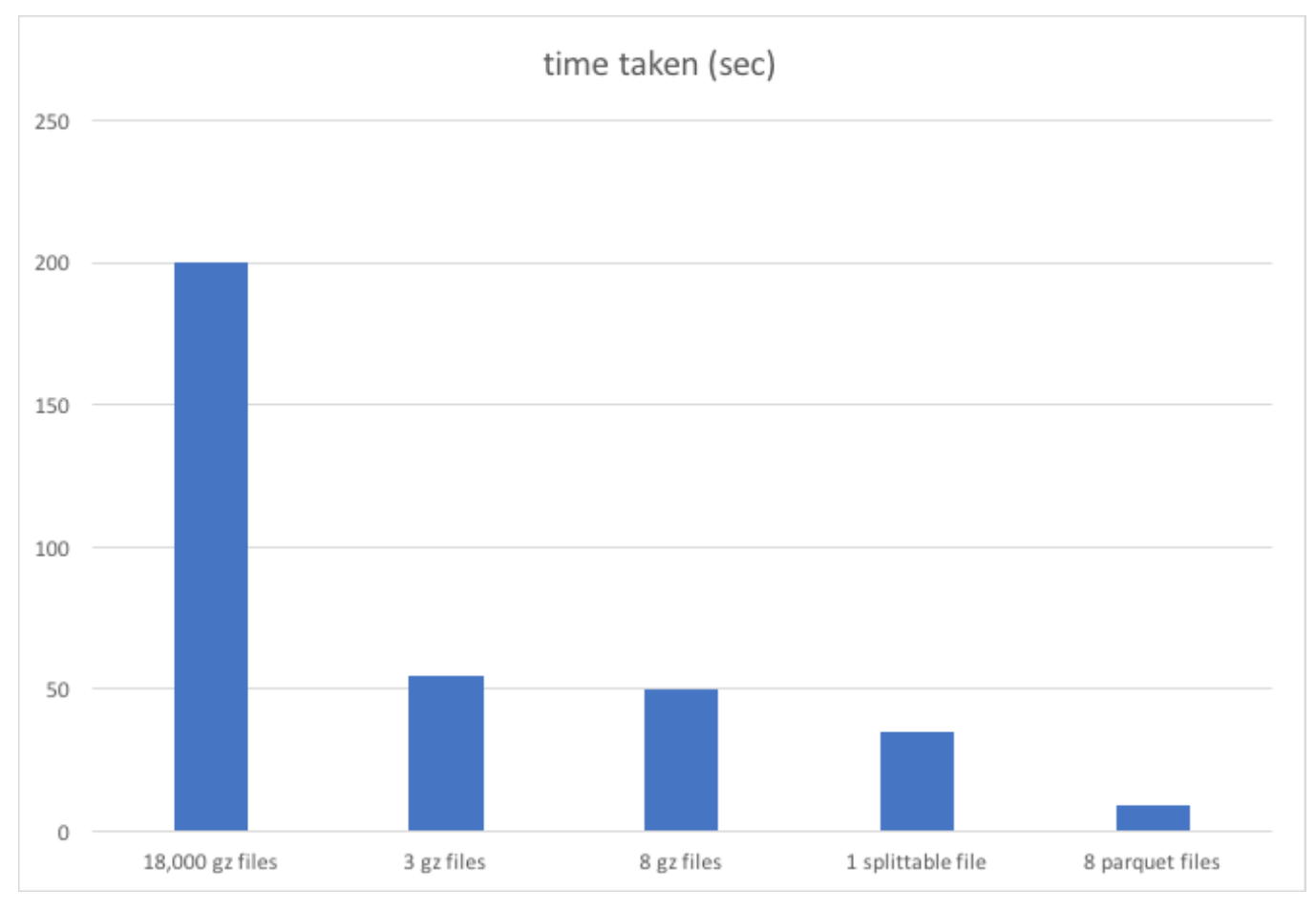
If the code and explanation doesn’t have you convinced, see this chart which shows the outsized performance boost by using Parquet over
armies of tiny files. This chart is only looking at one hour’s worth of data and on local HDFS instead of S3, so it actually makes the small files
look better than they should. I expect a minimum of a 20x speedup by using optimal file formats.
For the exceptionally interested parties, here is the chunk of spark code that I believe handles collecting the meta data on input files:
https://github.com/apache/spark/blob/master/sql/core/src/main/scala/org/apache/spark/sql/execution/datasources/DataSource.scala#L380 – note how it’s not parallelized, asynchronous or otherwise optimized for peak performance involving tiny files.
this is you explained in terms of s3, what if there is location in hdfs that contains 1000 files of each 2 MB size.
csv files.
Reading through spark.
how many partitions should be there in respective dataframe created.
Please explain with example….using spark-scala…
While 1,000 2MB files on HDFS is not optimal due to being smaller than HDFS block size (64 or 128MB default), it shouldn’t be a big issue. The number of partitions should be between 8 and 1,000 depending on compression (if any). If using DataFrames or Datasets, you can do df.rdd.getNumPartitions to get the total number of partitions.
What if you already have an army of tiny json files (~10KB) in S3. Is there a way to preliminarily convert those files to parquet in S3?
Mohammad, if you already have an army of tiny files, you can do a batch conversion to parquet by reading in chunks of the data set (e.g. 1 hour partition at a time of a 24 hour day)
Oh do you mean to simulate a stream? That’s a pretty cool idea. My approach, which worked, is a little different. I used python’s zipfile module. Since I had the files stored locally I was able to use python to append json files together to make bigger files of at least 128MB and at most 1GB. So far it seems to have worked. I went from having ~2M mini json files to 28 appropriately-sized json files (~300MB each). Your blog was super helpful, thanks!