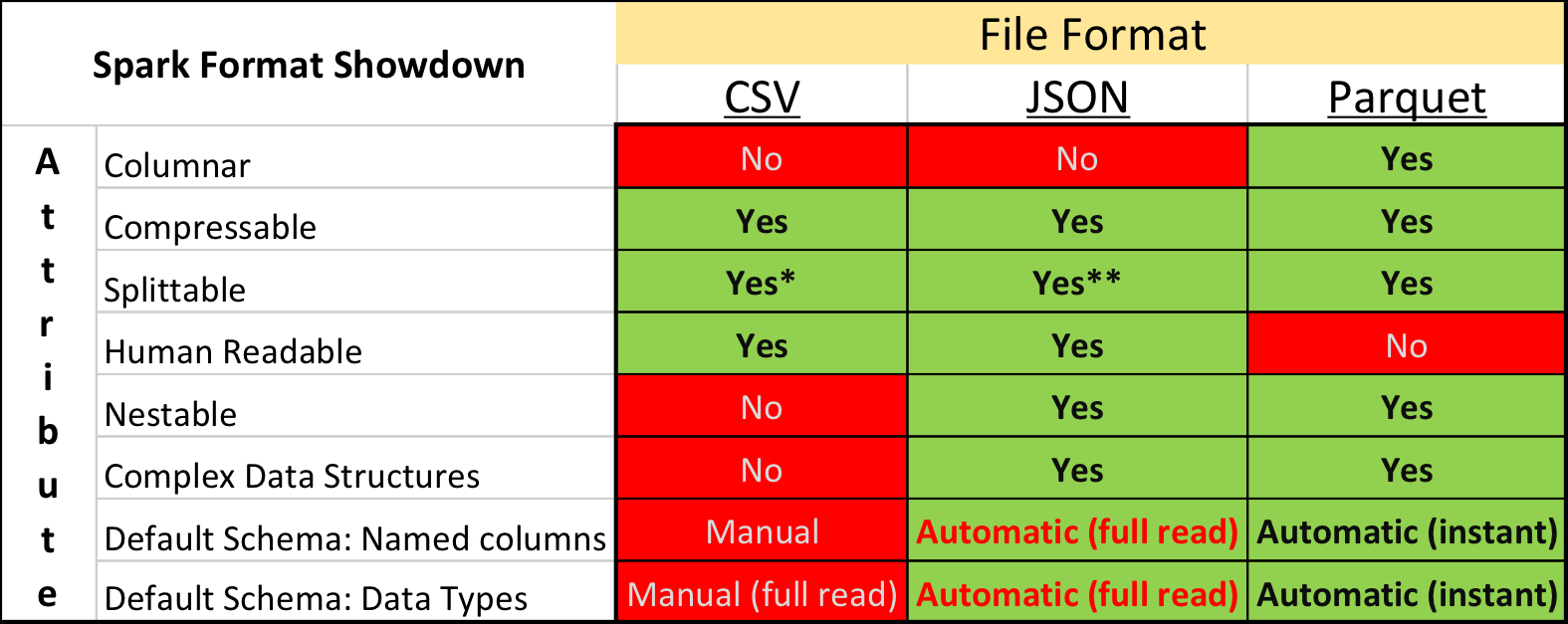
Apache Spark supports many different data sources, such as the ubiquitous Comma Separated Value (CSV) format and web API friendly JavaScript Object Notation (JSON) format. A common format used primarily for big data analytical purposes is Apache Parquet. Parquet is a fast columnar data format that you can read more about in two of my other posts: Real Time Big Data analytics: Parquet (and Spark) + bonus and Tips for using Apache Parquet with Spark 2.x
In this post we’re going to cover the attributes of using these 3 formats (CSV, JSON and Parquet) with Apache Spark.
Splittable (definition): Spark likes to split 1 single input file into multiple chunks (partitions to be precise) so that it [Spark] can work on many partitions at one time (re: concurrently).
* CSV is splittable when it is a raw, uncompressed file or using a splittable compression format such as BZIP2 or LZO (note: LZO needs to be indexed to be splittable!)
** JSON has the same conditions about splittability when compressed as CSV with one extra difference. When “wholeFile” option is set to true (re: SPARK-18352), JSON is NOT splittable.
CSV should generally be the fastest to write, JSON the easiest for a human to understand and Parquet the fastest to read.
CSV is the defacto standard of a lot of data and for fair reasons; it’s [relatively] easy to comprehend for both users and computers and made more accessible via Microsoft Excel.
JSON is the standard for communicating on the web. APIs and websites are constantly communicating using JSON because of its usability properties such as well-defined schemas.
Parquet is optimized for the Write Once Read Many (WORM) paradigm. It’s slow to write, but incredibly fast to read, especially when you’re only accessing a subset of the total columns. For use cases requiring operating on entire rows of data, a format like CSV, JSON or even AVRO should be used.
Code examples and explanations
CSV
Generic column names | all string types | lazily evaluated
scala> val df = spark.read.option("sep", "\t").csv("data.csv")
scala> df.printSchema
root
|-- _c0: string (nullable = true)
|-- _c1: string (nullable = true)
|-- _c2: string (nullable = true)
|-- _c3: string (nullable = true)
Header-defined column names | all string types | lazily evaluated
scala> val df = spark.read.option("sep", "\t").option("header","true").csv("data.csv")
scala> df.printSchema
root
|-- guid: string (nullable = true)
|-- date: string (nullable = true)
|-- alphanum: string (nullable = true)
|-- name: string (nullable = true)
Header-defined column names | inferred types | EAGERLY evaluated (!!!)
scala> val df = spark.read.option("sep", "\t").option("header","true").option("inferSchema","true").csv("data.csv")
scala> df.printSchema
root
|-- guid: string (nullable = true)
|-- date: timestamp (nullable = true)
|-- alphanum: string (nullable = true)
|-- name: string (nullable = true)
The eager evaluation of this version is critical to understand. In order to determine with certainty the proper data types to assign to each column, Spark has to READ AND PARSE THE ENTIRE DATASET. This can be a very high cost, especially when the number of files/rows/columns is large. It also does no processing while it’s inferring the schema, so if you thought it would be running your actual transformation code while it’s inferring the schema, sorry, it won’t. Spark has to therefore read your file(s) TWICE instead of ONCE.
JSON
Named columns | inferred types | EAGERLY evaluated
scala> val df = spark.read.json("data.json")
scala> df.printSchema
root
|-- alphanum: string (nullable = true)
|-- epoch_date: long (nullable = true)
|-- guid: string (nullable = true)
|-- name: string (nullable = true)
Like the eagerly evaluated (for schema inferencing) CSV above, JSON files are eagerly evaluated.
Parquet
Named Columns | Defined types | lazily evaluated
scala> val df = spark.read.parquet("data.parquet")
scala> df.printSchema
root
|-- alphanum: string (nullable = true)
|-- date: long (nullable = true)
|-- guid: string (nullable = true)
|-- name: string (nullable = true)
Unlike CSV and JSON, Parquet files are binary files that contain meta data about their contents, so without needing to read/parse the content of the file(s), Spark can just rely on the header/meta data inherent to Parquet to determine column names and data types.